What is Minerva?
Minerva is a suite of software tools for interpreting and interacting with complex images, organized around a guided analysis approach. The software enables fast sharing of large image data that is stored on Amazon S3 and viewed using a zoomable image viewer implemented using OpenSeadragon, making it ideal for integration into multi-omic browsers for data dissemination of tissue atlases. Check out the Minerva Wiki to learn more about the software and for news.
The Minerva suite includes Minerva Story, a narrative, web-based image viewer, and Minerva Author, a tool that enables the user to easily create a guided analysis, or story. Individuals with minimal coding experience can use Minerva Author to create a guided analysis of an image, which is then shared and visualized using Minerva Story. Guided analyses of multiplexed cyclic multiplex immunofluorescence (CyCIF), immunohistochemistry (IHC), and H&E images created by multiple lab members can be found on the data page and in the Featured Stories section below.
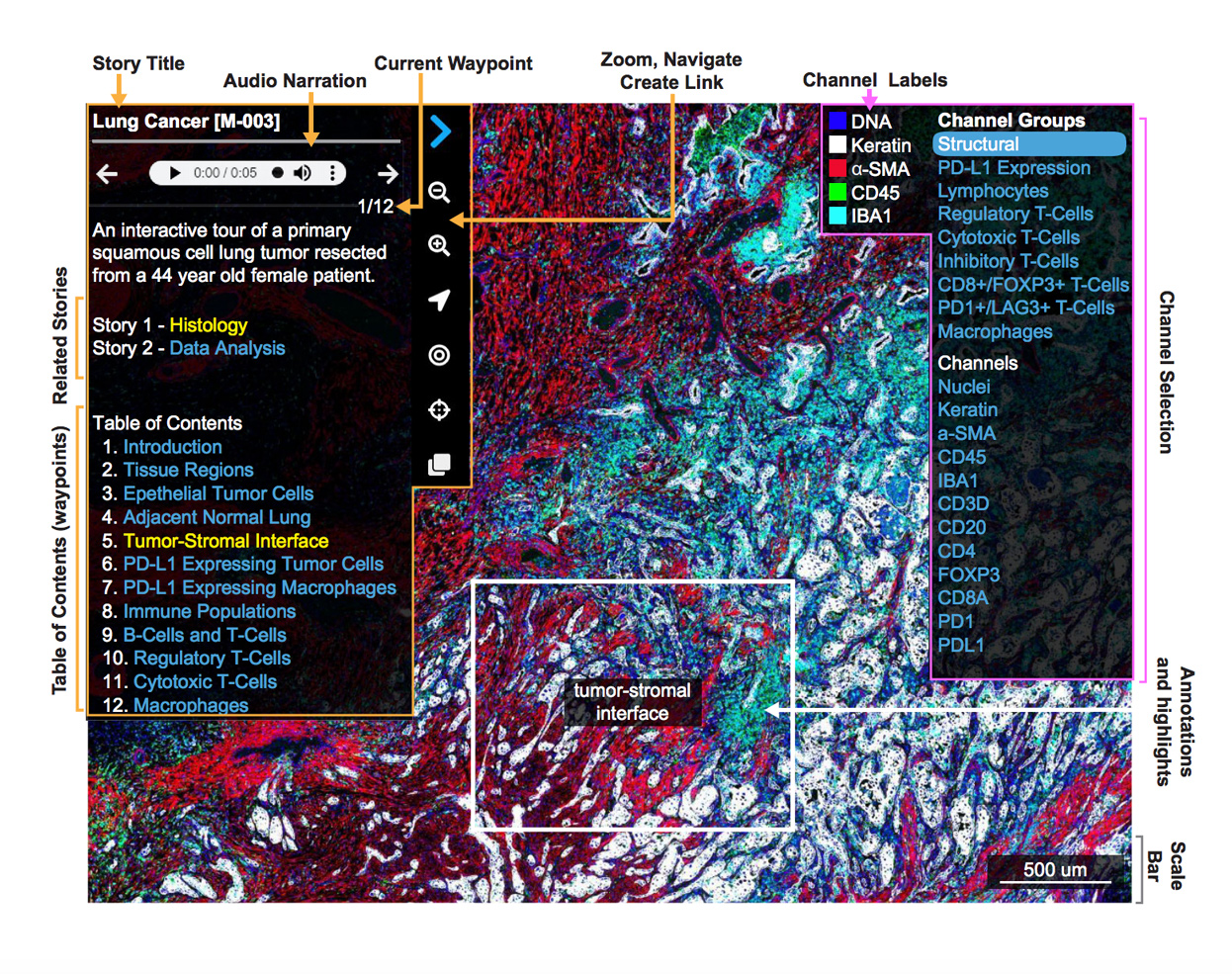
Featured Stories
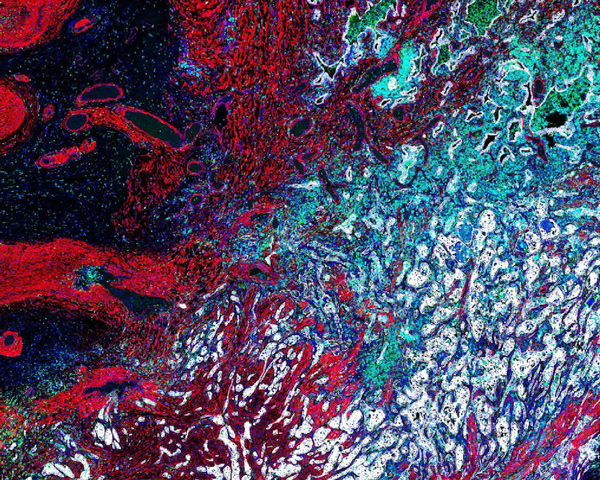
Lung Adenocarcinoma (Histology)
Explore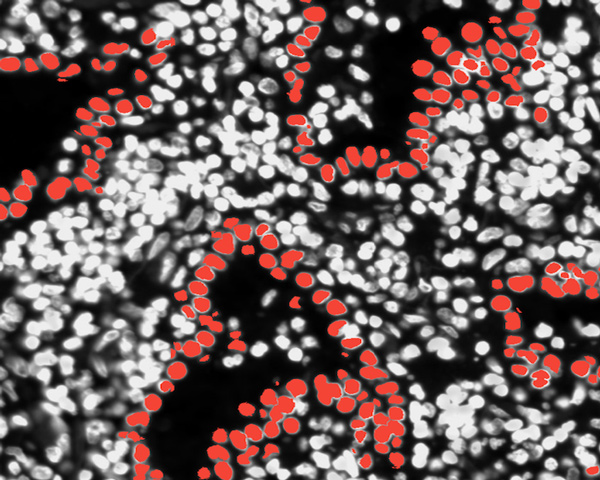
Lung Adenocarcinoma (Data Analysis)
Explore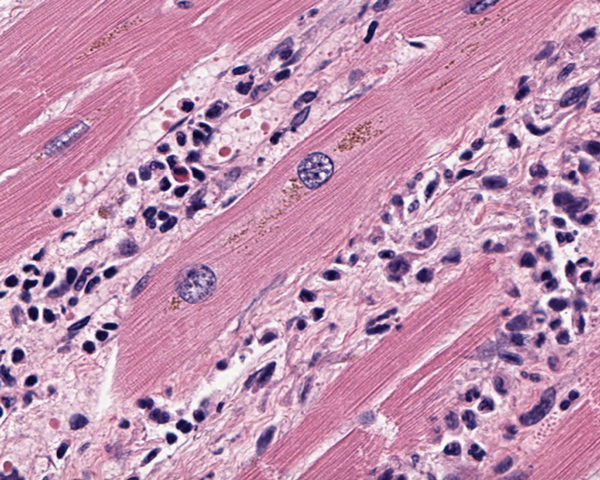
Multiple Myocardial Infarcts
Explore
Colorectal Adenocarcinoma
Explore
Melanoma Exceptional Responder
Explore
Melanoma
Explore
CNS Manifestations of Neurofibromatosis
Explore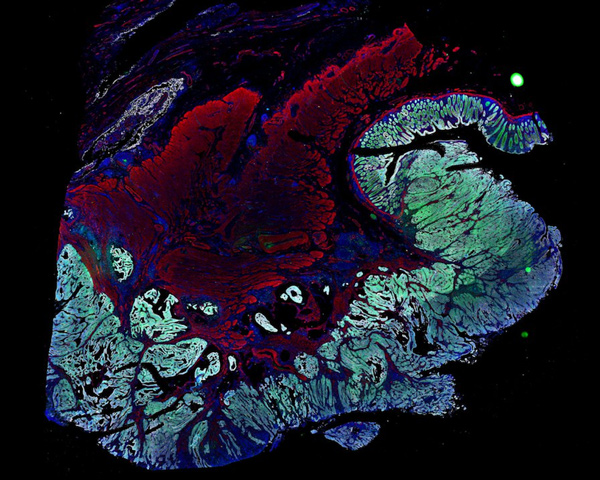
Colorectal Adenocarcinoma (3D Stack)
Explore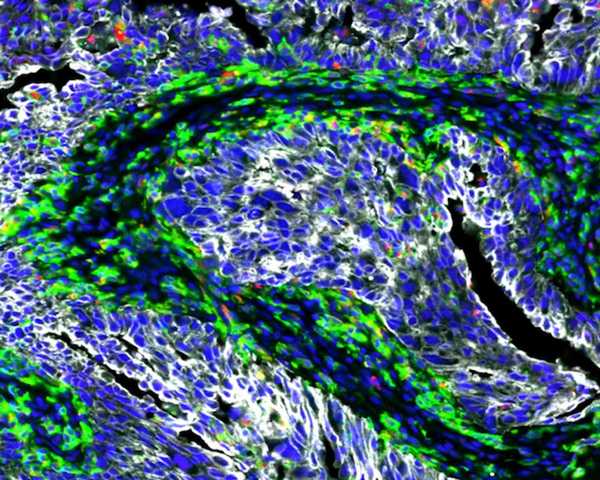
BRCA Mut v. WT Breast Cancer
ExploreHow does Minerva work?
Minerva follows a client-server model for delivering content. Images in OME-TIFF format are imported into Minerva Author where a user sets image settings and annotations. Minerva author then renders image pyramids and a configuration file that is read by Minerva Story to deliver the content to clients.
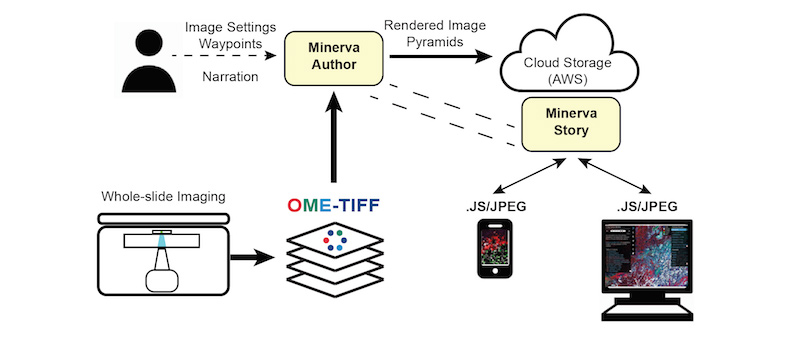
How do I implement Minerva?
Check out the Minerva Wiki for detailed user instructions, which walk the user through downloading and running Minerva Author, importing data, and creating channel groups to author a story. After authoring your story, you will deploy Minerva Story by cloning the repository on GitHub, adding your content to the repository, and deploying your story locally or via GitHub. We have chosen to deploy via GitHub and store data on Amazon S3 for universal sharing. All source code for Minerva is available on Minerva Github.
- Minerva Wiki https://github.com/labsyspharm/minerva-story/wiki
- Minerva Source Code https://github.com/labsyspharm/minerva-story
Publications
Rashid R, Chen YA, Hoffer J, Muhlich JL, Lin JR, Krueger R, Pfister H, Mitchell R, Santagata S, and Sorger PK. Interpretative guides for interacting with tissue atlas and digital pathology data using the Minerva browser. BioRxiv. (2020) https://doi.org/10.1101/2020.03.27.001834.
Hoffer J, Rashid R, Muhlich JL, Chen Y-A, Russell DPW, Ruokonen J, Krueger R, Pfister H, Santagata S, Sorger PK. Minerva: a light-weight, narrative image browser for multiplexed tissue images. J Open Source Software. 2020 Oct 15;5(54):2579.
Development Chart
| Concept | Req. Review | Development | Documentation | Stage | Rollout | Publication | |
|---|---|---|---|---|---|---|---|
| Minerva Story |  |
 |
 |
 |
beta | v1.0.0 | . |
| Minerva Author |  |
 |
 |
 |
beta | v1.2.0 | . |
| Minerva Cloud |  |
 |
 |
||||
| Minerva Analysis |  |
 |
 |
||||
| Minerva Atlas |  |
 |
 |

